JSON simulation language¶
For simple exchange of simulation descriptions a JSON format for the simulation was developed. The files for the code examples are available from https://github.com/matthiaskoenig/sbmlsim/tree/develop/docs/notebooks
import sbmlsim
print(sbmlsim.__version__)
0.1.0
Timecourse simulation¶
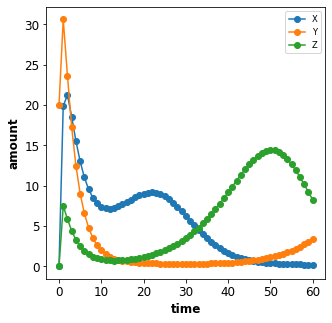
In the first example we create a simple timecourse simulation for the
repressilator model. The simulation starts at start ends at end
and has steps intervals (or steps+1 points).
from sbmlsim.timecourse import Timecourse, TimecourseSim
tcsim = TimecourseSim(Timecourse(start=0, end=60, steps=60))
We can convert the timecourse simulation into JSON via
json_str = tcsim.to_json()
print(json_str)
{
"timecourses": [
{
"normalized": false,
"start": 0,
"end": 60,
"steps": 60,
"changes": {},
"model_changes": {}
}
],
"selections": null,
"reset": true,
"time_offset": 0.0
}
By providing a file path we can store the definition as json file
tcsim.to_json('./json_examples/example_1.json')
We can now run the simulation with a given model, here with the
repressilator model and have a look at the numerical results.
from pathlib import Path
from sbmlsim.simulation_serial import SimulatorSerial as Simulator
simulator = Simulator("./models/repressilator.xml")
result = simulator.timecourses(tcsim)
print(result)
WARNING:pint.util:Redefining 'yr' (<class 'pint.definitions.UnitDefinition'>)
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [PX])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [PY])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [PZ])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [X])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [Y])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [Z])
WARNING:sbmlsim.units:DerivedUnit not found in UnitDefinitions: 1/(item)
WARNING:sbmlsim.units:DerivedUnit not found in UnitDefinitions: 1/(item)
WARNING:sbmlsim.units:DerivedUnit not found in UnitDefinitions: 1/(item)
<class 'sbmlsim.result.Result'>
DataFrames: 1
Shape: (61, 42, 1)
Size (bytes): 20496
# get timecourse data
result.mean.to_csv('./json_examples/example_1.tsv', index=False, sep='\t')
result.mean
WARNING:root:For a single simulation the mean returns the single simulation
| time | PX | PY | PZ | X | Y | Z | eff | n | KM | ... | Reaction10 | Reaction11 | Reaction12 | cell | [PX] | [PY] | [PZ] | [X] | [Y] | [Z] | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.0 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 20.000000 | 0.000000 | 20.0 | 2.0 | 40.0 | ... | 30.000000 | 30.000000 | 30.000000 | 1.0 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 20.000000 | 0.000000 |
| 1 | 1.0 | 81.440426 | 188.382020 | 42.641364 | 19.903426 | 30.615526 | 7.491014 | 20.0 | 2.0 | 40.0 | ... | 14.058083 | 5.854689 | 1.322932 | 1.0 | 81.440426 | 188.382020 | 42.641364 | 19.903426 | 30.615526 | 7.491014 |
| 2 | 2.0 | 218.538704 | 358.026782 | 84.588884 | 21.233519 | 23.608864 | 5.866252 | 20.0 | 2.0 | 40.0 | ... | 5.506924 | 1.001491 | 0.399478 | 1.0 | 218.538704 | 358.026782 | 84.588884 | 21.233519 | 23.608864 | 5.866252 |
| 3 | 3.0 | 337.622362 | 469.625864 | 113.054468 | 18.544097 | 17.234851 | 4.403620 | 20.0 | 2.0 | 40.0 | ... | 3.364327 | 0.444850 | 0.245856 | 1.0 | 337.622362 | 469.625864 | 113.054468 | 18.544097 | 17.234851 | 4.403620 |
| 4 | 4.0 | 428.938247 | 536.662776 | 131.034639 | 15.559775 | 12.482022 | 3.295798 | 20.0 | 2.0 | 40.0 | ... | 2.584705 | 0.288379 | 0.195576 | 1.0 | 428.938247 | 536.662776 | 131.034639 | 15.559775 | 12.482022 | 3.295798 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 56 | 56.0 | 218.684319 | 113.893055 | 1011.838390 | 0.257358 | 2.171448 | 11.909906 | 20.0 | 2.0 | 40.0 | ... | 0.076763 | 1.000240 | 3.320778 | 1.0 | 218.684319 | 113.893055 | 1011.838390 | 0.257358 | 2.171448 | 11.909906 |
| 57 | 57.0 | 205.724492 | 121.680149 | 1021.009829 | 0.246437 | 2.434108 | 11.063379 | 20.0 | 2.0 | 40.0 | ... | 0.075928 | 1.121739 | 2.952823 | 1.0 | 205.724492 | 121.680149 | 1021.009829 | 0.246437 | 2.434108 | 11.063379 |
| 58 | 58.0 | 193.568968 | 130.811333 | 1023.677923 | 0.238278 | 2.728748 | 10.156582 | 20.0 | 2.0 | 40.0 | ... | 0.075690 | 1.257369 | 2.592688 | 1.0 | 193.568968 | 130.811333 | 1023.677923 | 0.238278 | 2.728748 | 10.156582 |
| 59 | 59.0 | 182.181259 | 141.421225 | 1019.972101 | 0.232563 | 3.058478 | 9.218230 | 20.0 | 2.0 | 40.0 | ... | 0.076022 | 1.408327 | 2.250004 | 1.0 | 182.181259 | 141.421225 | 1019.972101 | 0.232563 | 3.058478 | 9.218230 |
| 60 | 60.0 | 171.525463 | 153.656610 | 1010.202008 | 0.229053 | 3.426504 | 8.275754 | 20.0 | 2.0 | 40.0 | ... | 0.076915 | 1.575792 | 1.932076 | 1.0 | 171.525463 | 153.656610 | 1010.202008 | 0.229053 | 3.426504 | 8.275754 |
61 rows × 42 columns
Now we create a small helper for plotting the results which we will reuse in the following examples.
%matplotlib inline
from sbmlsim import plotting_matplotlib
from matplotlib import pyplot as plt
def plot_repressilator_result(result):
df = result.mean
fig, (ax) = plt.subplots(nrows=1, ncols=1, figsize=(5, 5))
ax.plot(df.time, df.X, 'o-', label="X")
ax.plot(df.time, df.Y, 'o-', label="Y")
ax.plot(df.time, df.Z, 'o-', label="Z")
ax.set_xlabel("time")
ax.set_ylabel("amount")
ax.legend()
plt.show()
plot_repressilator_result(result)
Model changes¶
A simulation without changing anything in the model is a bit boring, so in the following we add changes to the model at the beginning of a simulation. Such changes can either be changes in the initial amount of a species, initial concentration of a species or parameter values.
Changes are defined via the changes field in a timecourse. The
referencing of model objects (species or parameters) works hereby via
the SId, i.e. the SBML identifiers used in the SBML model.
Parameter changes¶
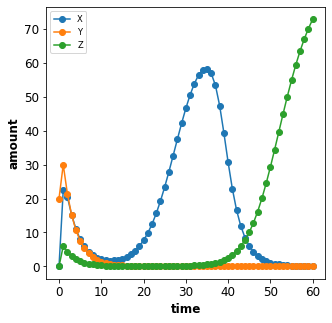
To change parameter values add the assignment of the change to the
changes dictionary. In the example the parameter with id n is
changed to 5 in the simulation by adding the changes {'n': 5} to
the Timecourse object.
ex_id = 2
simulator = Simulator("./models/repressilator.xml")
tcsim = TimecourseSim(
Timecourse(start=0, end=60, steps=60, changes={'n': 5})
)
result = simulator.timecourses(tcsim)
plot_repressilator_result(result)
tcsim.to_json(f'./json_examples/example_{ex_id}.json')
result.mean.to_csv(f'./json_examples/example_{ex_id}.tsv', index=False, sep='\t')
print(tcsim.to_json())
WARNING:pint.util:Redefining 'yr' (<class 'pint.definitions.UnitDefinition'>)
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [PX])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [PY])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [PZ])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [X])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [Y])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [Z])
WARNING:sbmlsim.units:DerivedUnit not found in UnitDefinitions: 1/(item)
WARNING:sbmlsim.units:DerivedUnit not found in UnitDefinitions: 1/(item)
WARNING:sbmlsim.units:DerivedUnit not found in UnitDefinitions: 1/(item)
WARNING:sbmlsim.timecourse:No units provided, assuming model units: n = 5 dimensionless
WARNING:root:For a single simulation the mean returns the single simulation
{
"timecourses": [
{
"normalized": true,
"start": 0,
"end": 60,
"steps": 60,
"changes": {
"n": {
"_magnitude": 5,
"_units": "dimensionless",
"_Quantity__used": false,
"_Quantity__handling": null
}
},
"model_changes": {}
}
],
"selections": null,
"reset": true,
"time_offset": 0.0
}
Initial amount changes¶
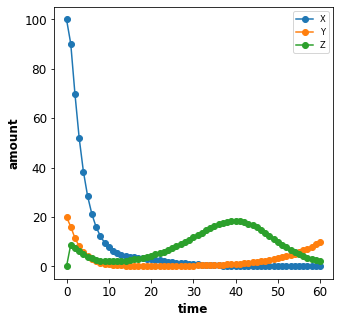
To change the initial amount of a species X to 100 we add the
corresponding changes {'X': 100} to the Timecourse.
ex_id = 3
simulator = Simulator("./models/repressilator.xml")
tcsim = TimecourseSim(
Timecourse(start=0, end=60, steps=60, changes={'X': 100})
)
result = simulator.timecourses(tcsim)
plot_repressilator_result(result)
tcsim.to_json(f'./json_examples/example_{ex_id}.json')
result.mean.to_csv(f'./json_examples/example_{ex_id}.tsv', index=False, sep='\t')
print(tcsim.to_json())
WARNING:pint.util:Redefining 'yr' (<class 'pint.definitions.UnitDefinition'>)
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [PX])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [PY])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [PZ])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [X])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [Y])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [Z])
WARNING:sbmlsim.units:DerivedUnit not found in UnitDefinitions: 1/(item)
WARNING:sbmlsim.units:DerivedUnit not found in UnitDefinitions: 1/(item)
WARNING:sbmlsim.units:DerivedUnit not found in UnitDefinitions: 1/(item)
WARNING:sbmlsim.timecourse:No units provided, assuming model units: X = 100 dimensionless
WARNING:root:For a single simulation the mean returns the single simulation
{
"timecourses": [
{
"normalized": true,
"start": 0,
"end": 60,
"steps": 60,
"changes": {
"X": {
"_magnitude": 100,
"_units": "dimensionless",
"_Quantity__used": false,
"_Quantity__handling": null
}
},
"model_changes": {}
}
],
"selections": null,
"reset": true,
"time_offset": 0.0
}
Initial concentration changes¶
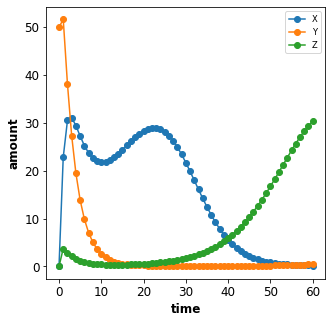
To change the initial concentration of a species Y to 50 in the
simulation we add the changes {'[Y]': 50} to the Timecourse. The
concentrations of species are referenced via the bracket syntax, i.e.
[Y] is the concentration of Y, whereas Y is the amount of
Y.
Note: For the repressilator model the volume in which the species
are located is 1.0, so that changes in amount correspond to changes
in concentration.
ex_id = 4
simulator = Simulator("./models/repressilator.xml")
tcsim = TimecourseSim(
Timecourse(start=0, end=60, steps=60, changes={'[Y]': 50})
)
result = simulator.timecourses(tcsim)
plot_repressilator_result(result)
tcsim.to_json(f'./json_examples/example_{ex_id}.json')
result.mean.to_csv(f'./json_examples/example_{ex_id}.tsv', index=False, sep='\t')
print(tcsim.to_json())
WARNING:pint.util:Redefining 'yr' (<class 'pint.definitions.UnitDefinition'>)
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [PX])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [PY])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [PZ])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [X])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [Y])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [Z])
WARNING:sbmlsim.units:DerivedUnit not found in UnitDefinitions: 1/(item)
WARNING:sbmlsim.units:DerivedUnit not found in UnitDefinitions: 1/(item)
WARNING:sbmlsim.units:DerivedUnit not found in UnitDefinitions: 1/(item)
WARNING:sbmlsim.timecourse:No units provided, assuming model units: [Y] = 50 dimensionless
WARNING:root:For a single simulation the mean returns the single simulation
{
"timecourses": [
{
"normalized": true,
"start": 0,
"end": 60,
"steps": 60,
"changes": {
"[Y]": {
"_magnitude": 50,
"_units": "dimensionless",
"_Quantity__used": false,
"_Quantity__handling": null
}
},
"model_changes": {}
}
],
"selections": null,
"reset": true,
"time_offset": 0.0
}
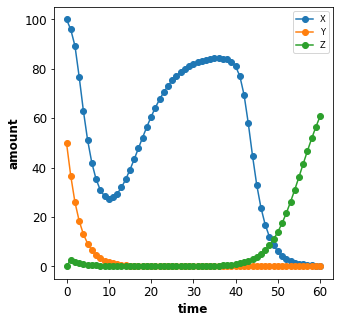
Combined changes¶
All these elementary changes can be combined and are all applied at the
beginning of the Timecourse. For instance to change the amount of
X to 100, the parameter n to 5 and the concentration of
[Y] to 50 use the changes
changes={'n': 5, 'X': 100, '[Y]: 50}
ex_id = 5
simulator = Simulator("./models/repressilator.xml")
tcsim = TimecourseSim(
Timecourse(start=0, end=60, steps=60, changes={'n': 5, 'X': 100, '[Y]': 50})
)
result = simulator.timecourses(tcsim)
plot_repressilator_result(result)
tcsim.to_json(f'./json_examples/example_{ex_id}.json')
result.mean.to_csv(f'./json_examples/example_{ex_id}.tsv', index=False, sep='\t')
print(tcsim.to_json())
WARNING:pint.util:Redefining 'yr' (<class 'pint.definitions.UnitDefinition'>)
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [PX])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [PY])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [PZ])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [X])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [Y])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [Z])
WARNING:sbmlsim.units:DerivedUnit not found in UnitDefinitions: 1/(item)
WARNING:sbmlsim.units:DerivedUnit not found in UnitDefinitions: 1/(item)
WARNING:sbmlsim.units:DerivedUnit not found in UnitDefinitions: 1/(item)
WARNING:sbmlsim.timecourse:No units provided, assuming model units: n = 5 dimensionless
WARNING:sbmlsim.timecourse:No units provided, assuming model units: X = 100 dimensionless
WARNING:sbmlsim.timecourse:No units provided, assuming model units: [Y] = 50 dimensionless
WARNING:root:For a single simulation the mean returns the single simulation
{
"timecourses": [
{
"normalized": true,
"start": 0,
"end": 60,
"steps": 60,
"changes": {
"n": {
"_magnitude": 5,
"_units": "dimensionless",
"_Quantity__used": false,
"_Quantity__handling": null
},
"X": {
"_magnitude": 100,
"_units": "dimensionless",
"_Quantity__used": false,
"_Quantity__handling": null
},
"[Y]": {
"_magnitude": 50,
"_units": "dimensionless",
"_Quantity__used": false,
"_Quantity__handling": null
}
},
"model_changes": {}
}
],
"selections": null,
"reset": true,
"time_offset": 0.0
}
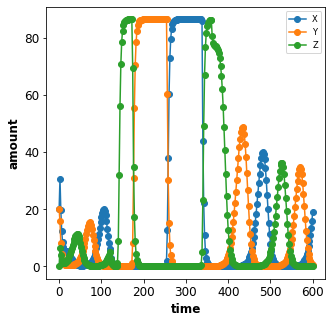
Combined timecourses¶
Multiple Timecourse objects can be combined to one large timecourse.
The results of the individual Timecourse are thereby concatenated.
The changes are always applied at the beginning of the individual
Timecourse simulations.
The model state is persistent in the multiple timecourses, i.e. the end state of the state variables of one Timecourse are the starting values of the next Timecourse (with exception of state variables affected by changes).
An example will demonstrate what is meant by this. The complete
timecourse simulation consists of 3 Timecourse parts:
start the first Timecourse with an initial amount of
X=20and simulate for 120 time stepsset
n=20in the model (while keeping the current state of all state variables) and continue simulating for 240 stepsset
n=2(this is the initial value of n) and continue simulating for another 240 steps
The result is a single timecourse simulation consisting of 3 timecourse parts.
ex_id = 6
simulator = Simulator("./models/repressilator.xml")
tcsim = TimecourseSim([
Timecourse(start=0, end=120, steps=60, changes={'X': 20}),
Timecourse(start=0, end=240, steps=120, changes={'n': 20}),
Timecourse(start=0, end=240, steps=120, changes={'n': 2}),
])
result = simulator.timecourses(tcsim)
plot_repressilator_result(result)
tcsim.to_json(f'./json_examples/example_{ex_id}.json')
result.mean.to_csv(f'./json_examples/example_{ex_id}.tsv', index=False, sep='\t')
print(tcsim.to_json())
WARNING:pint.util:Redefining 'yr' (<class 'pint.definitions.UnitDefinition'>)
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [PX])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [PY])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [PZ])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [X])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [Y])
WARNING:sbmlsim.units:substance or volume unit missing, impossible to determine concentration unit for [Z])
WARNING:sbmlsim.units:DerivedUnit not found in UnitDefinitions: 1/(item)
WARNING:sbmlsim.units:DerivedUnit not found in UnitDefinitions: 1/(item)
WARNING:sbmlsim.units:DerivedUnit not found in UnitDefinitions: 1/(item)
WARNING:sbmlsim.timecourse:No units provided, assuming model units: X = 20 dimensionless
WARNING:sbmlsim.timecourse:No units provided, assuming model units: n = 20 dimensionless
WARNING:sbmlsim.timecourse:No units provided, assuming model units: n = 2 dimensionless
WARNING:root:For a single simulation the mean returns the single simulation
{
"timecourses": [
{
"normalized": true,
"start": 0,
"end": 120,
"steps": 60,
"changes": {
"X": {
"_magnitude": 20,
"_units": "dimensionless",
"_Quantity__used": false,
"_Quantity__handling": null
}
},
"model_changes": {}
},
{
"normalized": true,
"start": 0,
"end": 240,
"steps": 120,
"changes": {
"n": {
"_magnitude": 20,
"_units": "dimensionless",
"_Quantity__used": false,
"_Quantity__handling": null
}
},
"model_changes": {}
},
{
"normalized": true,
"start": 0,
"end": 240,
"steps": 120,
"changes": {
"n": {
"_magnitude": 2,
"_units": "dimensionless",
"_Quantity__used": false,
"_Quantity__handling": null
}
},
"model_changes": {}
}
],
"selections": null,
"reset": true,
"time_offset": 0.0
}
Fields overview¶
The following fields are available for TimecourseSim:
selections: which columns should be stored in the outputreset: should the model be reset at the beginning of a TimecourseSim (default=True)
The following fields are available for Timecourse:
start: start time of integrationend: end time of integrationsteps: steps in the integration, the final output hassteps+1pointschanges: dictionary of changes applied at the beginning of timecourse simulationmodel_changes:depecratedcan be ignored